sbmlutils: Python utilities for SBML¶
sbmlutils is a collection of python utilities for working with SBML models
implemented on top of the libSBML python bindings.
Features among others
- HTML reports of SBML models
- helpers for model creation, manipulation, and annotation
- interpolation functions to add experimental data to models
- dynamic flux balance analysis (DFBA)
- file converters (XPP)
The project code is available from https://github.com/matthiaskoenig/sbmlutils.
Installation¶
pip install sbmlutilsSBML report¶
- create HTML reports of SBML files
- easy navigation and filtering
fbcandcompsupport
In [2]:
from sbmlutils.report import sbmlreport
sbmlreport.create_sbml_report('./examples/glucose/Hepatic_glucose_3.xml',
out_dir='./examples/glucose', validate=True)
-------------------------------------------------------------------------------- ./examples/glucose/Hepatic_glucose_3.xml valid : TRUE check time (ms) : 0.405 --------------------------------------------------------------------------------
SBML model creator¶
- helper functions for generation of SBML models
- constructors with all fields
- patterns (like generate
ParametersforAssignmentRules) fbcandcomphelpers- unit support
https://sbmlutils.readthedocs.io/en/latest/notebooks/modelcreator.html#Create-FBA-Model
Misc¶
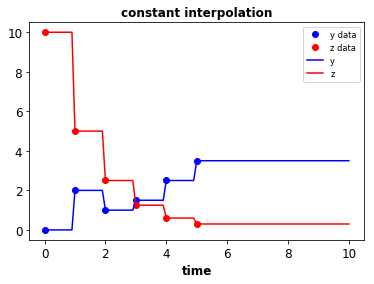
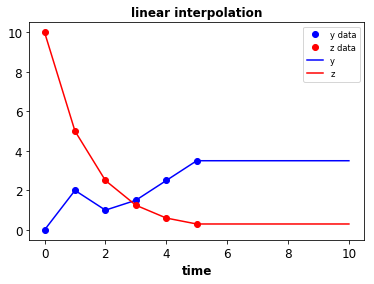
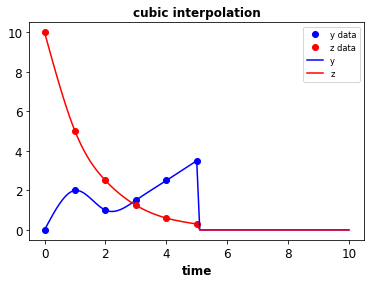
- Data to splines & piecewise functions
- Model annotation based on flat files and regular expressions
- converters (XPP -> SBML)
Dynamic FBA (DFBA)¶
- implementation of dynamic FBA based on comp and fbc
- model encoding & simulation
In [3]:
from __future__ import absolute_import, print_function
import os
import tempfile
import sbmlutils
from sbmlutils import dfba
from sbmlutils.dfba import utils
from sbmlutils.dfba.toy_wholecell import settings as toysettings
from sbmlutils.dfba.toy_wholecell import model_factory as toyfactory
from sbmlutils.dfba.toy_wholecell import simulate as toysimulate
test_dir = tempfile.mkdtemp()
# create toy model
toyfactory.create_model(test_dir)
sbml_path = os.path.join(utils.versioned_directory(test_dir, toyfactory.version),
toysettings.top_file)
print(sbml_path)
Create directory: /tmp/tmp3sm90cd5/v12 -------------------------------------------------------------------------------- /tmp/tmp3sm90cd5/v12/toy_wholecell_fba.xml valid : TRUE check time (ms) : 0.011 -------------------------------------------------------------------------------- -------------------------------------------------------------------------------- /tmp/tmp3sm90cd5/v12/toy_wholecell_bounds.xml valid : TRUE check time (ms) : 0.012 -------------------------------------------------------------------------------- -------------------------------------------------------------------------------- /tmp/tmp3sm90cd5/v12/toy_wholecell_update.xml valid : TRUE check time (ms) : 0.007 -------------------------------------------------------------------------------- -------------------------------------------------------------------------------- /tmp/tmp3sm90cd5/v12/toy_wholecell_top.xml valid : TRUE check time (ms) : 0.195 -------------------------------------------------------------------------------- Flattened model written to /tmp/tmp3sm90cd5/v12/toy_wholecell_flattened.xml /tmp/tmp3sm90cd5/v12/toy_wholecell_fba.xml /tmp/tmp3sm90cd5/v12/toy_wholecell_bounds.xml /tmp/tmp3sm90cd5/v12/toy_wholecell_update.xml /tmp/tmp3sm90cd5/v12/toy_wholecell_top.xml /tmp/tmp3sm90cd5/v12/toy_wholecell_flattened.xml /tmp/tmp3sm90cd5/v12/toy_wholecell_top.xml
In [4]:
from IPython.display import display, HTML
# simulate
dfs = toysimulate.simulate_toy(sbml_path, test_dir, dts=[1.0], figures=False)
display(dfs[0].head(10))
toysimulate.print_species(dfs=dfs)
toysimulate.print_fluxes(dfs=dfs)
Simulation time : 0.255 [s] Total time : 0.339 [s] Overhead time : 0.084 [s] (24.8 %) * DFBA Solution is UNIQUE *
| time | [A] | [C] | [D] | [dummy_S] | [fba__A] | [fba__B1] | [fba__B2] | [fba__C] | EX_A | ... | fba__ub_default | fba__zero | k_R4 | lb_EX_A | lb_EX_C | pEX_A | pEX_C | ub_EX_A | ub_EX_C | ub_R1 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.0 | 0.0 | 10.000000 | 0.000000 | 0.000000 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | -1.000000 | ... | 1000.0 | 0.0 | 0.1 | -100.000000 | 0.000000 | -1.000000 | 1.000000 | 1000.0 | 1000.0 | 1.000000 |
| 1.0 | 1.0 | 9.000000 | 0.951626 | 0.048374 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | -0.818731 | ... | 1000.0 | 0.0 | 0.1 | -90.000000 | -9.516258 | -0.818731 | 0.818731 | 1000.0 | 1000.0 | 0.818731 |
| 2.0 | 2.0 | 8.181269 | 1.640192 | 0.178539 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | -0.670320 | ... | 1000.0 | 0.0 | 0.1 | -81.812693 | -16.401919 | -0.670320 | 0.670320 | 1000.0 | 1000.0 | 0.670320 |
| 3.0 | 3.0 | 7.510949 | 2.122001 | 0.367049 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | -0.548811 | ... | 1000.0 | 0.0 | 0.1 | -75.109491 | -21.220014 | -0.548811 | 0.548811 | 1000.0 | 1000.0 | 0.548811 |
| 4.0 | 4.0 | 6.962138 | 2.442333 | 0.595529 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | -0.449323 | ... | 1000.0 | 0.0 | 0.1 | -69.621378 | -24.423330 | -0.449323 | 0.449323 | 1000.0 | 1000.0 | 0.449323 |
| 5.0 | 5.0 | 6.512814 | 2.637502 | 0.849683 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | -0.367873 | ... | 1000.0 | 0.0 | 0.1 | -65.128144 | -26.375023 | -0.367873 | 0.367873 | 1000.0 | 1000.0 | 0.367873 |
| 6.0 | 6.0 | 6.144941 | 2.736589 | 1.118470 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | -0.301187 | ... | 1000.0 | 0.0 | 0.1 | -61.449411 | -27.365889 | -0.301187 | 0.301187 | 1000.0 | 1000.0 | 0.301187 |
| 7.0 | 7.0 | 5.843754 | 2.762785 | 1.393460 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | -0.246589 | ... | 1000.0 | 0.0 | 0.1 | -58.437541 | -27.627855 | -0.246589 | 0.246589 | 1000.0 | 1000.0 | 0.246589 |
| 8.0 | 8.0 | 5.597165 | 2.734532 | 1.668303 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | -0.201888 | ... | 1000.0 | 0.0 | 0.1 | -55.971651 | -27.345320 | -0.201888 | 0.201888 | 1000.0 | 1000.0 | 0.201888 |
| 9.0 | 9.0 | 5.395277 | 2.666429 | 1.938295 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | -0.165291 | ... | 1000.0 | 0.0 | 0.1 | -53.952767 | -26.664287 | -0.165291 | 0.165291 | 1000.0 | 1000.0 | 0.165291 |
10 rows × 30 columns
In [ ]:
